Metabolic Flux Analysis with 13C-Labeling Experiments
To deeply understand the cells' metabolism is indispensable when aiming at an increased productivity under industrially relevant conditions by targeted genetic manipulation. A wide range of omics-tools comprising genomic and metagenomic sequencing, transcriptome, proteome, and metabolome profiling are nowadays available. Due to post-transcriptional regulation and enzymatic activities, cellular processes might not be sufficiently reflected in these omics profiles. The entirety of all metabolic flux rates, the fluxome, is the physiologically most relevant description of metabolic activity.
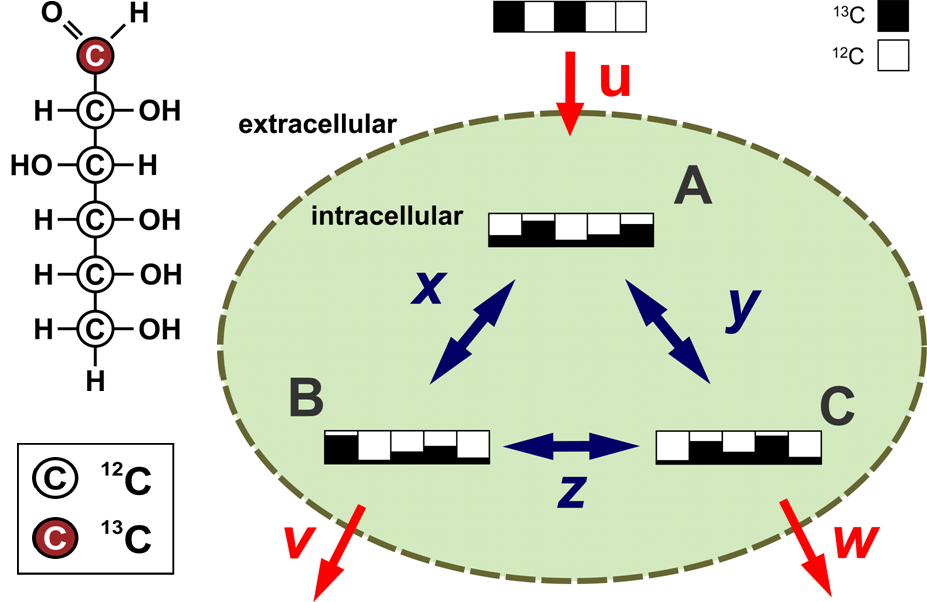
Intracellular metabolic fluxes, however, can not be directly assessed in vivo but have to be inferred from measured quantities. Currently the most reliable method for determining these unobservable reaction rates in a metabolically steady state is metabolic flux analysis with 13C-labeling experiments (13C-MFA): A defined 13C-labeled substrate is incorporated into the carbon backbone of a wide range of metabolites, the metabolome, either through exchange or by synthesis. The distribution of labeled carbon traversing along metabolic pathways generates a characteristic imprint of labeling patterns whose mass signature is observed by mass spectrometry (MS). All fluxes can be determined on an absolute scale if physiological fluxes in and out of cells are available.
As part of an integrative data evaluation approach including many scientific disciplines (genomics, biochemistry, process engineering, chemical analytics, and computer-based modeling), 13C-MFA has become one of the major tools in Metabolic Engineering and Systems Biology. 13C-MFA is a versatile model-based tool is typically used to support the following three key issues:
- Strain characterization:
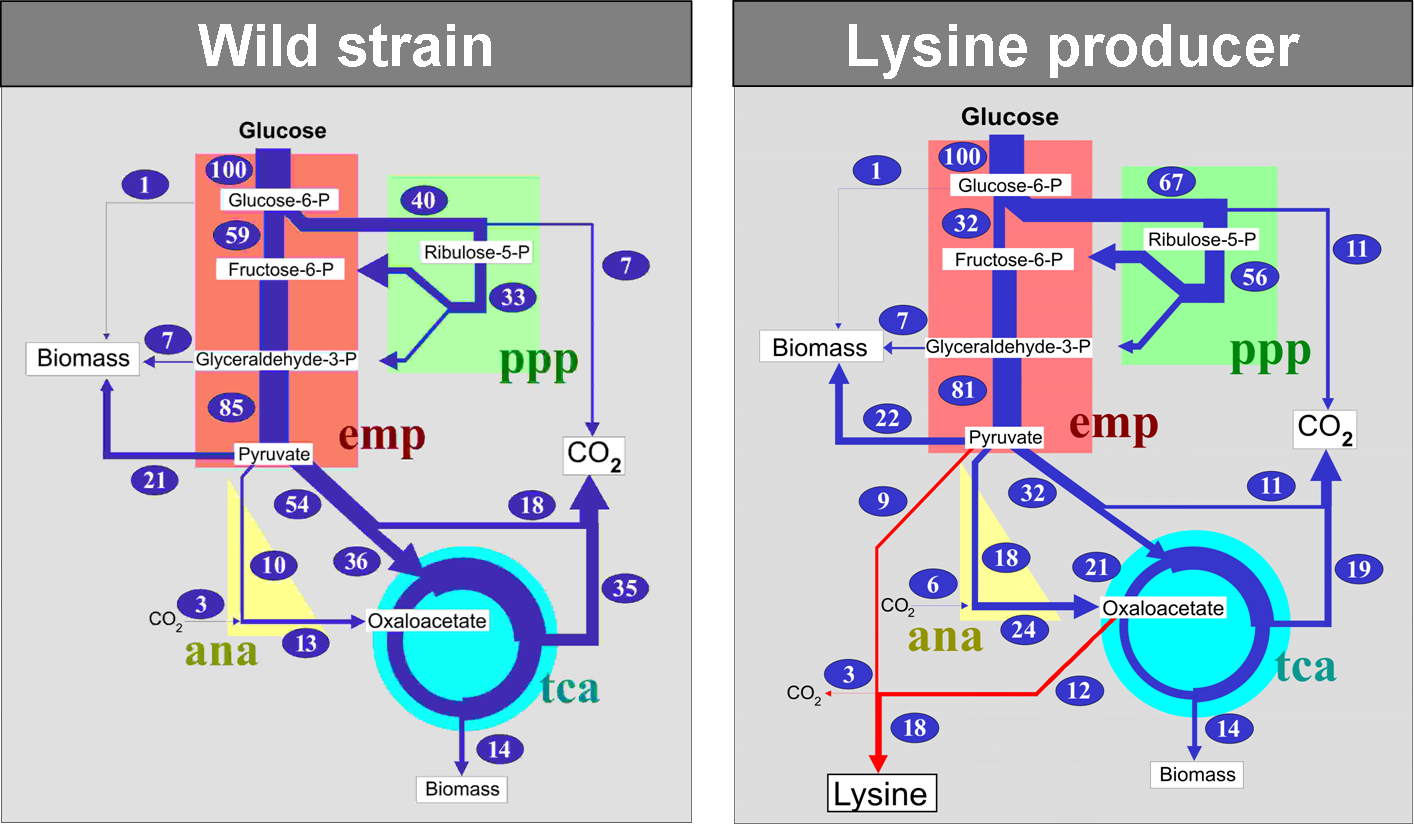
Generating and improving insights into metabolic pathway activity by comparing flux phenotypes under different environmental conditions and physiological states as well as for a variety of carbon sources. - Metabolic engineering targets:
Identifying pathway bottlenecks in order to maximize metabolite synthesis. This, in turn, can help to derive decisive hypotheses for promising gen targets. - Hypothesis validation:
Gene function manipulations can be verified on metabolome and fluxome level.
Summarizing, 13C-MFA facilitates direct and comprehensive quantitative insights into the active metabolic network and is nowadays applied to a diversity of organisms. In particular, functional understanding enzymatic reactions in these organisms would pave the way for the development of novel, more efficient design of strains as well as biotechnological processes.
icons by famfamfam.com last change: 29.09.2023 | IBT Webmaster | Print


