13CFLUX2:
 The High-Performance
Simulator for
The High-Performance
Simulator for
13C-based Metabolic Flux Analysis
State-of-the-Art Algorithms for Ultimate Universality
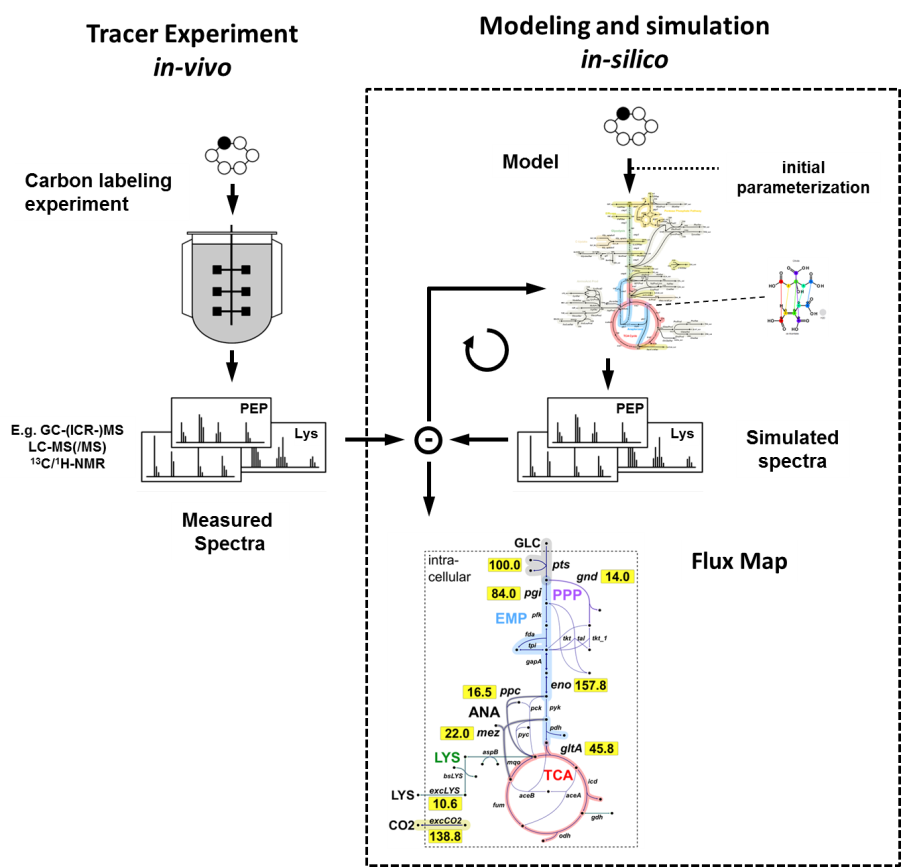 Metabolic fluxes are the final endpoint of all co-operating actions in
the complex cellular network of genes, transcripts, proteins and
metabolites. In vivo fluxes, however, are not directly observable and
have to be inferred by data from labeling experiments and the use of
mathematical models. 13C-based metabolic flux analysis emerged as the
state-of-the-art technique in the field of fluxomics.
Metabolic fluxes are the final endpoint of all co-operating actions in
the complex cellular network of genes, transcripts, proteins and
metabolites. In vivo fluxes, however, are not directly observable and
have to be inferred by data from labeling experiments and the use of
mathematical models. 13C-based metabolic flux analysis emerged as the
state-of-the-art technique in the field of fluxomics.
The
high-performance simulator 13CFLUX2 is a software suite of
applications for the detailed quantification of intracellular (quasi)
steady-state fluxes .
The typical evaluation
workflow with 13CFLUX2 consists of several steps:
- custom network modeling and measurement specification
- simulation of isotope labeling states
- parameter estimation (globalized strategies line multi-start heuristics)
- statistical quality analysis (non-linear and linearized)
- experimental design
Major Features
- Highly efficient implementation of Cumomer and Elementary
Metabolite Unit (EMU) simulation algorithms
(operating with arbitrary precision if needed) - Specification of arbitrary measurement s (e.g. LC-MS/MS, 13C-NMR)
- Bindings for powerful optimization frameworks (e.g. IPOPT, NAG-C)
- FluxML, a standardized XML-based file format the formulation of 13C modles
Look and Feel
13CFLUX2 consists of multiple applications providing a command-line interface. Analysis results are stored as hdf5 and csv files facilitating an easy post-processing with, e.g., Matlab™. The visualization tool Omix provides an easy-to-use graphical modeling front- and backend for 13CFLUX2.Benchmark
S. cerevisiae network with 313 metabolites and 359 reactions is supplemented with a full set of carbon atom transitions (longest c-atom chain 57). Realistic flux distributions and measurement specifications are taken from literature. Simulation runs for predicting measured labeling patterns ("forward problem") took approx. 200 ms on a (old-fashioned) notebook.How to Cite?
If you publish or publicly disclose the results of using the simulator 13CFLUX2, please acknowledge 13CFLUX2 in your publications by citing the following reference:13CFLUX2 - High-Performance Software Suite for 13C-Metabolic Flux Analysis
Bioinformatics 29(1): 143-5, 2013
doi: 10.1093/bioinformatics/bts646
Download Paper
Availability and Licensing
A standalone version of 13CFLUX2 is available under a commercial license. Please contact us for the details.For academic usage in scientific research projects (e.g. for universities and non-profit research institutes) 13CFLUX2 will be available free of charge under a non-commercial license. This academic license is not restricted in functionality but requires an active internet connection and SSL-encrypted file exchange between the working machine and our license server (i.e., a HTTPS connection is mandatory).
System Requirements
13CFLUX2 has been developed on LINUX / UNIX operating systems and tested to run under the following LINUX distributions:- Ubuntu 64bit LTS
icons by famfamfam.com last change: 29.09.2023 | IBT Webmaster | Print


